User Tools
Sidebar
DIMEDR - Disparate Metabolomics Data Reassembler
 In the past decade, the field of LC-MS based metabolomics has transformed from an obscure specialty into a major “-omics” platform for studying metabolic processes and biomolecular characterization. However, as a whole the field is still very fractured, as the nature of the instrumentation and of the information produced by the platform essentially creates incompatible “islands” of datasets. This lack of data coherency results in the inability to accumulate a critical mass of metabolomics data that has enabled other –omics platforms to make impactful discoveries and meaningful advances.
In the past decade, the field of LC-MS based metabolomics has transformed from an obscure specialty into a major “-omics” platform for studying metabolic processes and biomolecular characterization. However, as a whole the field is still very fractured, as the nature of the instrumentation and of the information produced by the platform essentially creates incompatible “islands” of datasets. This lack of data coherency results in the inability to accumulate a critical mass of metabolomics data that has enabled other –omics platforms to make impactful discoveries and meaningful advances.
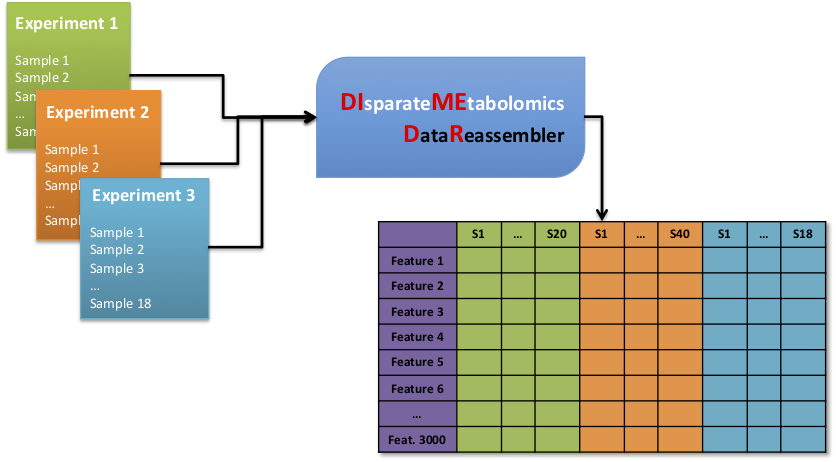
As such, we have developed a novel algorithm, called Disparate Metabolomics Data Reassembler (DIMEDR), which attempts to bridge the inconsistencies between incongruent LC-MS metabolomics datasets of the same biological sample type. A single “primary” dataset is postprocessed via traditional means of peak identification, alignment, and grouping. DIMEDR utilizes this primary dataset as a progenitor template by which data from subsequent disparate datasets are reassembled and integrated into a unified framework that maximizes spectral feature similarity across all samples. This is accomplished by a novel procedure for universal retention time correction and comparison via identification of ubiquitous features in the initial primary dataset, which are subsequently utilized as endogenous internal standards during integration.
For demonstration purposes, two human and two mouse urine metabolomics datasets from four unrelated studies acquired over 4 years were unified via DIMEDR, which enabled meaningful analysis across otherwise incomparable and unrelated datasets.
The program can be downloaded here, along with details installation instructions and a getting started guide.
For suggestions, inquiries, and comments, please contact Tytus Mak (tytus.mak@nist.gov).
