User Tools
Sidebar
Table of Contents
NIST MSQC Pipeline
Attention: Support for NIST MSQC Pipeline is discontinued
Software for Monitoring LC-MS Performance
This program was written to systematically evaluate analytical performance of a common discovery-based proteomics platform by monitoring selected output from a liquid chromatography -mass spectrometry (LC-MS/MS) system. The software was developed to help researchers identify sources of variations due to analytical problems. The idea behind the software is that if analytical variations can be minimized using defined mixtures and metrics, variations due to biological differences in complex samples can be more confidently identified.
NIST MSQC expects that the MS/MS spectra are produced from analysis of a tryptic digest of a protein mixture. However, future releases will allow any set of MS/MS data files, such as those from analysis of metabolites or other small molecules and can be identified by a spectral library search, to be used. Typically, data files might come from routine analysis of a QC standard over time but may be from different instruments in the same or different laboratories. Analysis is carried out by calculating and comparing a defined set of data metrics from one or more set of MS data files. The use of this program does not require any new data acquisition; it has been designed to be run post-data acquisition and is therefore suitable for examining older data files for historical purposes. However, best practice for this software would be iterations of controlled runs of a sample, followed data analysis, and correction of any problems identified by fluctuations in key metrics.
This software contains many component software applications which are controlled by a single Perl program. The software is intended to be run as a “pipeline.” That is, processing starts from RAW mass spectrometry data files and is passed through many programs, where output from the previous application is required by the next. It is therefore important that the program be allowed to run in its entirety. Progress of the pipeline is reported by the software and a reasonable attempt has been made to catch common errors and provide suitable remedies.
Important Note on Data Formats: Use of this version is limited to analysis of data acquired on Thermo Scientific ion traps (e.g., LTQ, LTQ-Orbitrap, FT-LTQ) and Agilent QTOF mass spectrometers generating MS/MS spectra by CID or HCD. Future releases may allow comparison of data files generated on mass spectrometers produced by other manufacturers provided sources of accessible data formats can be identified.
Additional Requirements
- Program runs under Microsoft© Windows™ operation system (XP or later versions)
- XCalibur™ (preferably the same version used on the data acquisition computer) or Thermo Scientific MSFileReader
- Perl (freely available from (freely available from http://www.activestate.com/activeperl)
Download and Installation Instruction
Attention: Support for NIST MSQC Pipeline is discontinued
- 1. Download the NIST MSQC Pipeline software appropriate for your Windows operation system. If you are in doubt, select 32-bit version.
- 2. Extract the NIST MSQC Pipeline zip archive in a working directory on your hard drive (e.g., C:\projects\NIST_MSQC_Pipeline).
- 3. One or more search library/database packs is required. These should be relevant to the sample you are using. For example, if your QC sample is S. cerevesiae, you will need to download and extract the yeast.zip archive in the 'libs' directory of your installation. Other library packs can be provided by request. Download one or more peptide library packs (bsa.zip, ecoli.zip, sigmaups1.zip, yeast.zip), or, minimally, an MSPepSearch library in 'NIST' format from peptide.nist.gov and move the extracted folder and/or files to your libs sub-directory.
Operating Instructions
The main window of the program is divided into three tabs:
More detailed description for each tab is followed.
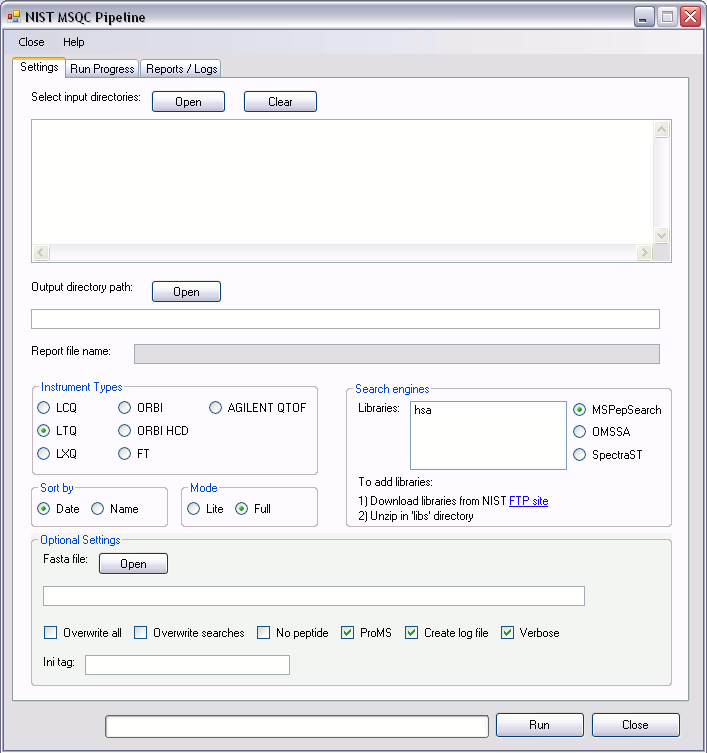
Settings
- Input directories: To run the program we need to provide input data file for processing. User can select at least one directory containing Thermo .RAW files or previously generated MGF+MS1 files. If you wish to use previously generated MGF+MS1 files, these MUST have been produced by a recent version of ReAdW4Mascot2.
- Output directory selection: User should select output directory (folder) where all intermediate output files are to be written (Figure 3). NIST MSQC will write output files from all input directories to this path. By default NIST MSQC's working directory is 'out_dirs'. It is recommended to create new output directories within 'out_dir'. User can also choose any other directory.
- Report file name: The name of the report file is automatically generated by the program using the format output_directory_name_report.msqc (Figure 5). This tab-delimited, text file contains all of the calculated metrics values for all runs and series. If this file name has been previously used, NIST MSQC will “increment” the file name. For example, instead of overwriting
test_report.msqc, the filetest_report——–1.msqcwill be created instead. - Instrument types: It is used to specify the instrument model used to generate the data. If ORBI or FT is specified, precursor tolerance will be reduced and monoisotopic precursor masses will be used. Additionally, ReAdW4Mascot2 will attempt to correct any precursor mass miscalls made by XCalibur™ by re-evaluating the MS1 isotopic envelope for the sampled ion.
- Search engines: The three options are MSPepSearch, OMSSA or SpectraST. These are used to specify the search engine identifying the MS/MS spectra. OMSSA is a sequence search engine and will only search BLAST-formatted databases. These are provided in the bundles (bsa.zip, ecoli.zip, sigmaups1.zip, yeast.zip). The other two search engines require MS/MS mass spectral libraries.
- Sort: The option is used to order the runs as columns in the final output file. If names of files give a better ordering (“ASCII-betically”) within a series, name should be specified. The default is sort by date.
- Mode: In the Full running mode additional peptide and protein-level analysis will be performed. NOTE: Information generated in the output by specifying this option is experimental and no documentation is available.
- Optional Settings
- Fasta File: If not specified, NIST MSQC will automatically look for a fasta file in the 'libs' directory of the library selected in the 'Libraries' box. FASTA files are ONLY used during protein mapping by nistms_metrics (i.e., if mode was set to full) and are NOT required for general use. Additionally, if MSPepSearch was used to search multiple libraries AND mode was set to full, you will need to create a FASTA file containing sequences found in all libraries specified.
- Overwrite all: If checked, all processing on RAW files will be repeated and any output files will be overwritten. Useful if converter has been updated, for example.
- Overwrite searches: If checked, searches will re-run and older output files (TSV or pepXML) will be overwritten. Peak list converter (i.e., ReAdW4Mascot2) will not re-run.
- No peptide: If checked, only a spectral library search engine is allowed and FASTA option is ignored. Useful if searching a library of metabolite MS/MS spectra.
- ProMS: If checked, results from ProMS, a NIST - developed MS1 data analysis program, will be used instead of values calculated by ReAdW4Mascot2. ProMS requires MS1 profile spectra mzXML format, so these (large) files will additionally be generated during data conversions. ProMS's XIC methods give more consistent results especially for high-resolution MS1 data.
- Create log file: If checked, output from the pipeline will be sent to a log file. These files can be viewed in the “Summary/Logs” tab with the extension .LOG.
- Verbose: If checked, more information on the progress of the run will be displayed while the pipeline is executing.
- Ini tag: Name of tag in scripts\ms.ini file, (e.g, test}. This option allows manual editing of a section of the ms.ini file. This may be used to re-run an analysis after changing the ordering of files or by grouping them as series. To group a set of runs by series, insert SERIES between FILE links. In order to use this option, edit the value between the braces in ms.ini and save the file. NOTE: Editing this file incorrectly may cause problems; use caution when editing the file. Additionally, no checking of other arguments in the ms.ini section is currently done or are tags checked for duplication. If the specified tag appears more than once, the first one will be used.
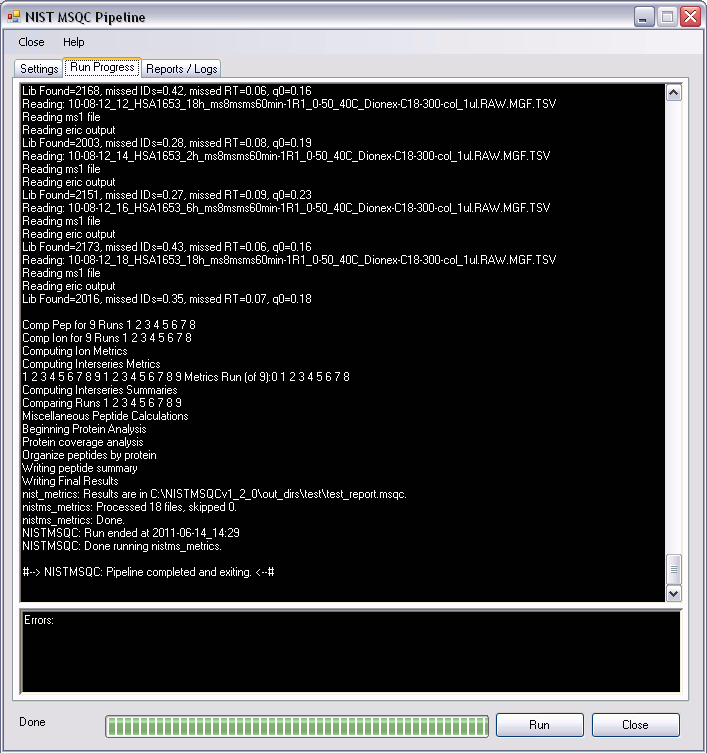
Run Progress
- While processing the data NIST MSQC Pipeline will display output information in the output box found in Run Progress tab. By checking the “Verbose” option on “Settings” tab the user can see more information on run progress
- Errors - Any errors encountered during the processing of a run will be displayed in the error box found in “Run Progress” tab.
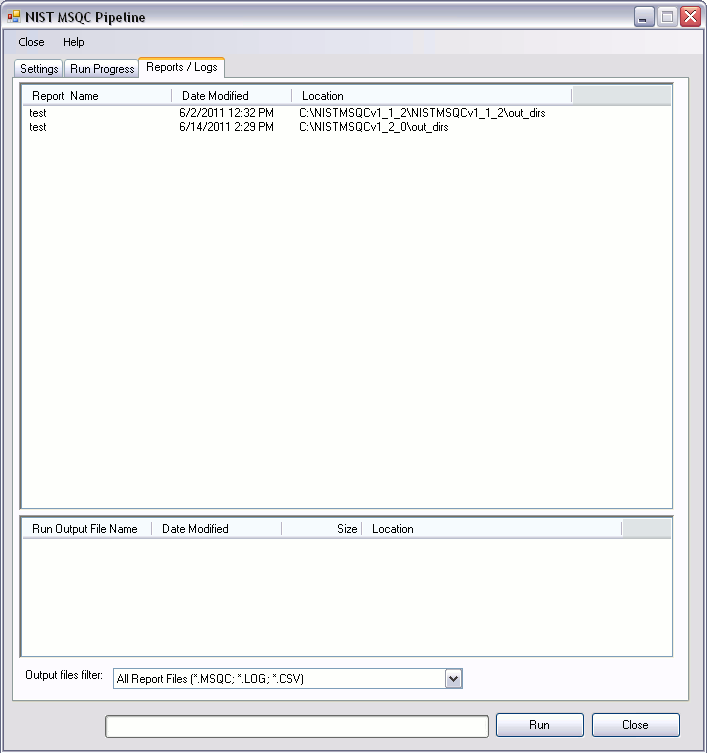
Reports / Logs
- Every run generates a number of different logs and reports. These reports can be viewed in “Reports / Logs” tab. The top list displays the name of the report. The report name is identical to the name of the folder selected in output directory path. Clicking on a report name will populate the bottom list which contains list of files generated during a run
- The list of report files can be filtered to view only the desired files.
Additional Information
This document is intended to provide instructions on how to run the NIST MSQC software on your computer. The metrics and benchmark values have been reported in two publications (PubMed IDs: 19837981, 19858499). If you have questions about the metrics, please refer to these documents first.
For more background information, you may also wish to visit http://proteomics.cancer.gov/
Attention: Support for NIST MSQC Pipeline is discontinued